PhylOTU
:: DESCRIPTION
PhylOTU identifies microbial operational taxonomic units (OTUs) from metagenomic data.
::DEVELOPER
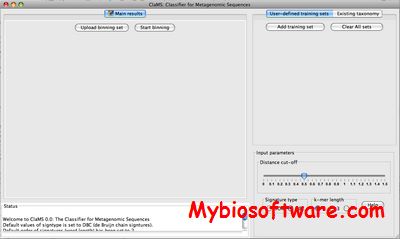
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows/ MacOsX
- Perl
- BIoPerl
- C++ COmpiler
:: DOWNLOAD
:: MORE INFORMATION
Citation
PLoS Comput Biol. 2011 Jan 20;7(1):e1001061. doi: 10.1371/journal.pcbi.1001061.
PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data.
Sharpton TJ1, Riesenfeld SJ, Kembel SW, Ladau J, O’Dwyer JP, Green JL, Eisen JA, Pollard KS.