MICE 1.4
:: DESCRIPTION
MICE (Mouse Information and Classification Entity) is a program aimed at facilitating the monitoring of animals in their facility. It consists of a virtual facility in which scientists can perform all the tasks done in the real world (i.e., receiving animals, breeding, etc…). Each animal is recorded with all associated information (birth date, cage number, ID number, tail analysis number, parents, genetic status, genetic background and more), allowing for reliable tracking. Animals can be identified, grouped, sorted, moved…, according to any parameter of interest to the scientist, including associated comments. Crossings are automatically processed by the program, which determines the new genetic background, generation number, cage location and due date.
MICE reminds the user when births are expected, and entering the newborn animals only requires a few clicks (of the mouse!). The genealogy of each animal can be determined in two different ways, including a visual tree from which each ancestor’s information can be retrieved.
::DEVELOPER
P. Pognonec
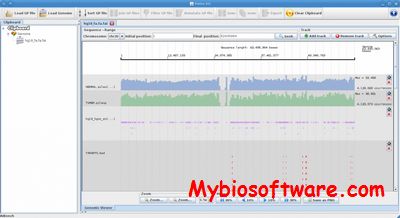
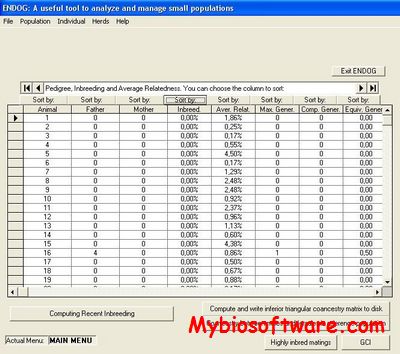
:: SCREENSHOTS

:: REQUIREMENTS
:: DOWNLOAD
 MICE for Win ; for Mac
MICE for Win ; for Mac
:: MORE INFORMATION
Citation:
MICE, a program to track and monitor animals in animal facilities. BMC Genetics 2001 Mar;2(1):4
Any comment and/or proposal concerning this application is welcome! Please contact P. Pognonec for additional information.
 BioLegend CD Posters / BioLegend Tools for iPad
BioLegend CD Posters / BioLegend Tools for iPad