Endog 4.8
:: DESCRIPTION
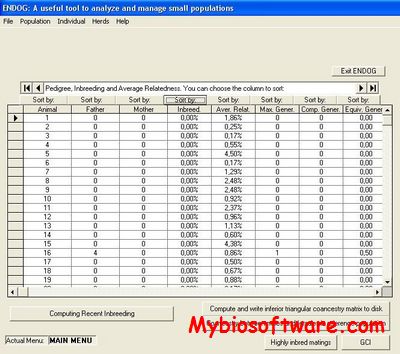
ENDOG‘s Primary functions are the computation of the individual inbreeding (F) (Wright, 1931) and the average relatedness (AR) (Gutiérrez et al., 2003; Goyache et al., 2003) coefficients. Additionally, users can compute with ENDOG useful parameters in population genetics such as that described for Biochard et al. (1997) for the number of ancestors explaining genetic variability or those proposed by Robertson (1953) and Vassallo et al. (1996) for the genetic importance of the herds. ENDOG also can compute F statistics (Wright, 1978) from genealogical information following Caballero and Toro (2000; 2002). Moreover, the present version of ENDOG calculates effective population size following different methodologies including regression approaches and particularly the recently proposed realized effective population size from individual increase in inbreeding (Gutiérrez et al., 2008), modified to account for avoidance of self-fertilization (Gutiérrez et al., 2009).
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
A note on ENDOG: a computer program for analysing pedigree information.
Gutiérrez JP, Goyache F.
J Anim Breed Genet. 2005 Jun;122(3):172-6.