SPC Proteomics Tools 060107
:: DESCRIPTION
SPC Proteomics Tools corom contains Seattle Proteome Center (SPC) – Proteomics Tools.
::DEVELOPER
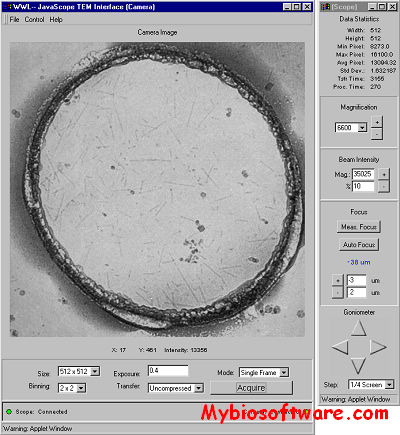
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows / Mac OsX
:: DOWNLOAD
:: MORE INFORMATION
this CD image contains an old version of the SPC tools. Please see the TPP wiki page for the most updated information.