Scipion 3.0
:: DESCRIPTION
Scipion is an image processing framework to obtain 3D models of macromolecular complexes using Electron Microscopy (3DEM). It integrates several software packages and presents an unified interface for both biologists and developers.
::DEVELOPER
Biocomputing Unit – CNB
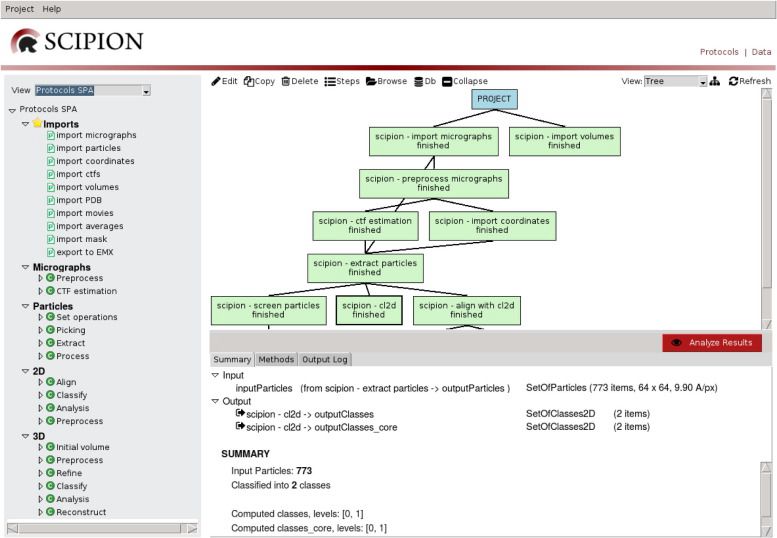
:: SCREENSHOTS
:: REQUIREMENTS
- Linux
- Xmipp
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Scipion: a software framework toward integration, reproducibility and validation in 3D Electron Microscopy.
de la Rosa-Trevín JM, Quintana A, Del Cano L, Zaldívar A, Foche I, Gutiérrez J, Gómez-Blanco J, Burguet-Castell J, Cuenca-Alba J, Abrishami V, Vargas J, Otón J, Sharov G, Vilas JL, Navas J, Conesa P, Kazemi M, Marabini R, Sorzano CO, Carazo JM.
J Struct Biol. 2016 Apr 20. pii: S1047-8477(16)30079-X. doi: 10.1016/j.jsb.2016.04.010
A statistical approach to the initial volume problem in Single Particle Analysis by Electron Microscopy.
Sorzano CO, Vargas J, de la Rosa-Trevín JM, Otón J, álvarez-Cabrera AL, Abrishami V, Sesmero E, Marabini R, Carazo JM.
J Struct Biol. 2015 Mar;189(3):213-9. doi: 10.1016/j.jsb.2015.01.009.