GoSurfer 2.0
:: DESCRIPTION
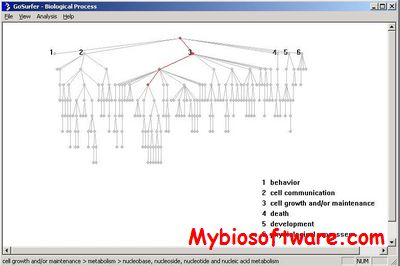
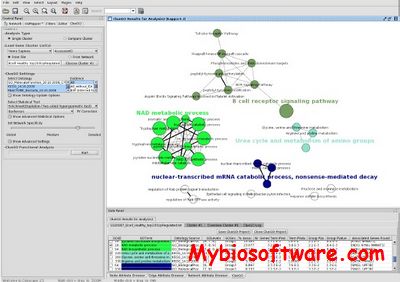
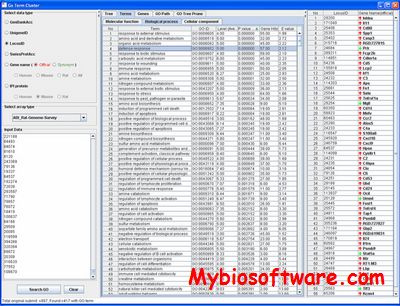
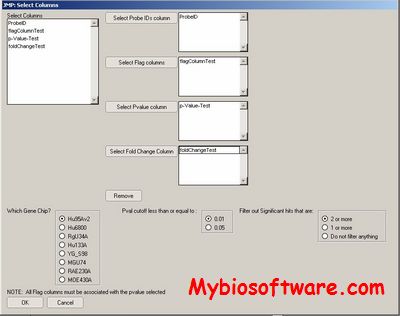
GoSurfer uses Gene Ontology (GO) information to analyze gene sets obtained from genome-wide computations or microarray analyses. GoSurfer is a graphical interactive data mining tool. It associates user input genes with GO terms and visualizes such GO terms as a hierarchical tree. Users can manipulate the tree output by various means, like setting heuristic thresholds or using statistical tests. Significantly important GO terms resulted from a statistical test can be highlighted. All related information are exportable either as texts or as graphics.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Zhong S, Storch F, Lipan O, Kao MJ, Weitz C, Wong WH.
GoSurfer: a graphical interactive tool for comparative analysis of large gene sets in Gene Ontology space.
Applied Bioinformatics 2004, 3(4): 1-5.