ARGminer
:: DESCRIPTION
ARGminer is an online resource for the inspection and curation of ARGs based on crowdsourcing as well as a platform to promote interaction and collaboration for the ARG scientific community.
::DEVELOPER
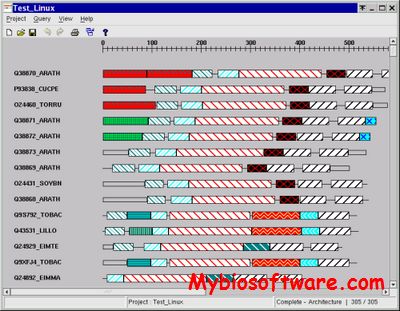
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
Arango-Argoty GA, Guron GKP, Garner E, Riquelme MV, Heath LS, Pruden A, Vikesland PJ, Zhang L.
ARGminer: a web platform for the crowdsourcing-based curation of antibiotic resistance genes.
Bioinformatics. 2020 May 1;36(9):2966-2973. doi: 10.1093/bioinformatics/btaa095. PMID: 32058567.
 NO
NO