mkdom/Xdom 2
:: DESCRIPTION
mkdom is the program we use routinely to build each new release of ProDom, Protein domain families automatically generated from protein databases. It relies on the assumption that the shortest amino acid sequence corresponds to a single domain, and may be used as a query to screen the database with the psi-blast program, in order to cluster homologous domains. For building ProDom, we run this program on the whole swissprot/trembl database, but it can be run on any set of protein sequences (as long as you have a fasta file).
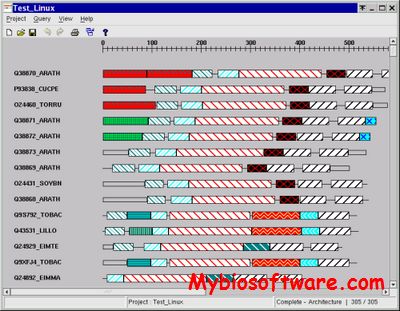
xdom is a graphical program which will help you to analyze domains detected by mkdom2, as it visualises all domain arrangements in the protein set.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Gouzy J., Corpet F. & Kahn D. (1999).
Whole genome protein domain analysis using a new method for domain clustering,
Computers and Chemistry. 23:333-340.
GOUZY J., EUGENE P. , GREENE E.A. , KAHN D. 1 and CORPET F.
XDOM, a graphical tool to analyse domain arrangements in any set of protein sequences
Comput Appl Biosci (1997) 13 (6): 601-608.