BioParishodhana
:: DESCRIPTION
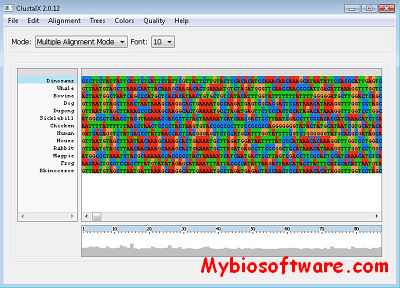
BioParishodhana integrates tools for similarity search, primer designing, and restriction enzyme digestion are required in almost all biological research.
::DEVELOPER
Institute of Bioinformatics and Applied Biotechnology, Bangalore, India,
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
Vangala RK, Singh L, Gupta RP.
BioParishodhana: A novel graphical interface integrating BLAST, ClustalW, primer3 and restriction digestion tools.
Bioinformation. 2012;8(13):639-43. doi: 10.6026/97320630008639. Epub 2012 Jul 6. PMID: 22829746; PMCID: PMC3400990.