Ergatis v2r18
:: DESCRIPTION
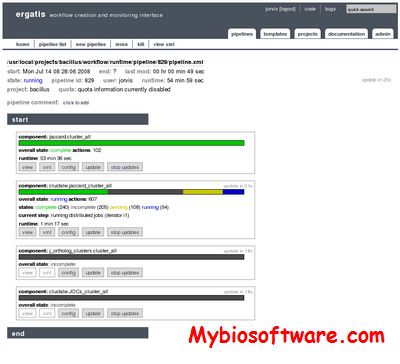
Ergatis is a web-based utility that is used to create, run, and monitor reusable computational analysis pipelines. It contains pre-built components for common bioinformatics analysis tasks. These components can be arranged graphically to form highly-configurable pipelines. Each analysis component supports multiple output formats, including the Bioinformatic Sequence Markup Language (BSML). The current implementation includes support for data loading into project databases following the CHADO schema, a highly normalized, community-supported schema for storage of biological annotation data.
::DEVELOPER
The Institute for Genome Sciences (IGS)
:: SCREENSHOTS
:: REQUIREMENTS
- Workflow Engine
- Sun Grid Engine
- Web server
- Perl
:: DOWNLOAD
 Ergatis
Ergatis
:: MORE INFORMATION
Citation
Orvis J, Crabtree J, Galens K, Gussman A, Inman JM, Lee E, Nampally S, Riley D, Sundaram JP, Felix V, Whitty B, Mahurkar A, Wortman J, White O, Angiuoli SV.
Ergatis: A web interface and scalable software system for bioinformatics workflows.
Bioinformatics. 2010 Jun 15;26(12).