MOIRAI 20140528
:: DESCRIPTION
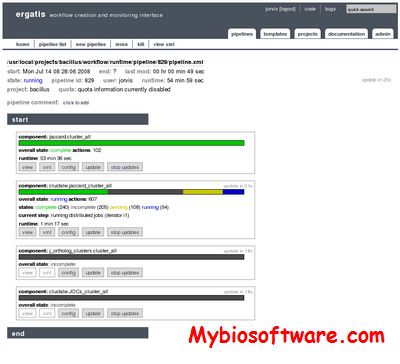
MOIRAI is a compact yet flexible workflow system designed to carry out the main steps in data processing and analysis of CAGE data. MOIRAI has a graphical interface allowing wet-lab researchers to create, modify and run analysis workflows. Embedded within the workflows are graphical quality control indicators allowing users assess data quality and to quickly spot potential problems.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- web2.0 client/server
- Java
- c++ server infrastructure
:: DOWNLOAD
:: MORE INFORMATION
Citation
MOIRAI: a compact workflow system for CAGE analysis.
Hasegawa A, Daub C, Carninci P, Hayashizaki Y, Lassmann T.
BMC Bioinformatics. 2014 May 16;15(1):144. doi: 10.1186/1471-2105-15-144.