APEX 1.1.0
:: DESCRIPTION
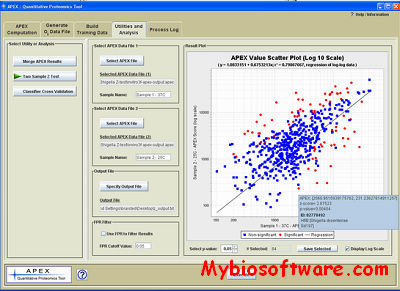
APEX (Absolute protein expression) Quantitative Proteomics Tool, is a free and open source Java implementation of the APEX technique for the quantitation of proteins based on standard LC- MS/MS proteomics data (Lu et al., Nature Biotech., 25(1):117-124, 2007). The underlying technique improves upon label free quantitation techniques such as MS/MS spectral counting. This technique uses machine learning algorithms to help correct for variable peptide detection related to certain peptide physicochemical properties which favor or hinder detection. The APEX tool provides a graphical user interface, an extensive integrated user help system, and a complete user’s manual. A sample data set and descriptive tutorial help to acquaint users with the operation of the tool. The APEX tool provides full support for all aspects of the APEX protein quantitation method and offers several utilities to help support downstream analysis. Please refer to the manual for a full description of the tool’s capabilities.
::DEVELOPER
the Marcotte Lab at University of Texas at Austin
:: SCREENSHOTS

:: REQUIREMENTS
- Linux / Windows / Mac OsX
- JAVA
:: DOWNLOAD
 APEX
APEX
:: MORE INFORMATION
Citation:
Peng Lu etc.
Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation,
Nature Biotechnology 25, 117 – 124 (2007)