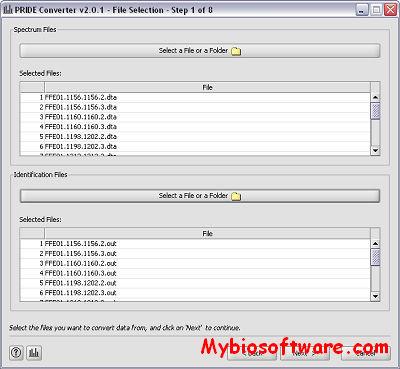
XML2RAF 1.02
:: DESCRIPTION
XML2RAF is a Perl software to create RAF maps from PDB XML files.The Protein Data Bank provides XML files that include a mapping between the PDB-format records SEQRES (representing the sequence of the molecule used in an experiment) and ATOM (representing the atoms experimentally observed). These XML files (along with the PDB’s chemical dictionary) also provide information on the original identity of most residues prior to any post-translational modifications, including cyclizations.
::DEVELOPER
Computational Genomics Research Group
:: SCREENSHOTS
N/A
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE.
The ASTRAL compendium in 2004.
Nucleic Acids Research 32:D189-D192 (2004).