MeV 4.9.0
:: DESCRIPTION
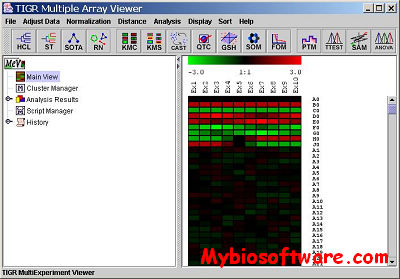
MeV is a desktop application for the analysis, visualization and data-mining of large-scale genomic data. It is a versatile microarray tool, incorporating sophisticated algorithms for clustering, visualization, classification, statistical analysis and biological theme discovery.
MeV allows the user to view processed microarray slide representations and identifies genes and expression patterns of interest. Slides can be viewed one at a time in detail or in groups for comparison purposes. A variety of normalization algorithms and clustering analyses allow the user flexibility in creating meaningful views of the expression data.
MeV generates informative and interrelated displays of expression and annotation data from single or multiple experiments. A huge array of alrogithms are included in MeV modules, and are available at a button-click, such as K-means clustering, Hierarchical clustering, t-Tests, Significance Analysis of Microarrays, Gene Set Enrichment Analysis, and EASE.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Mac OS X /Linux
- Java
:: DOWNLOAD
:: MORE INFORMATION
Referencing MeV
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003 Feb;34(2):374-8.