ICM-Browser 3.9-2b
:: DESCRIPTION
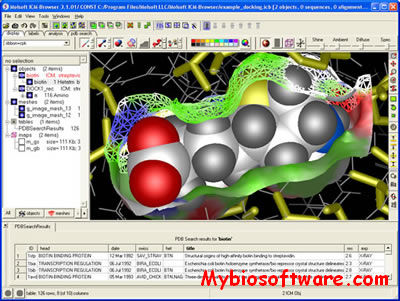
ICM-Browser provides a biologist or a chemist with direct access to the treasures of structural biology and protein families.It reads a variety of file formats directly from the database web-sites including: PDB, chemical, electron density maps, sequence and alignment files. ICM-Browser provides a rich professional molecular graphics environment with powerful representations of proteins, DNA and RNA, and multiple sequence alignments.
When you purchase the license ICM-Browser will become ICM-Browser-Pro.ICM-Browser-Pro is a high quality visualizer and annotator for three dimensional molecular structures, sequences, alignments, chemical spreadsheets and biological data. It allows you to read data from multiple file formats, annotate the data, and write multi-slide documents in a single small cross-platform file. ICM Browser Pro is well suited for creating, storing and sharing structural, biological and chemical information. The files can then be opened and viewed with the free ICM Browser.
::DEVELOPER
Molsoft LLC.
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
 ICM-Browser ; Manual
ICM-Browser ; Manual
:: MORE INFORMATION