VARNA 3-93
:: DESCRIPTION
VARNA (Viral Associated RNAs) is Java lightweight Applet dedicated to drawing the secondary structure of RNA. It is also a Swing component that can be very easily included in an existing Java code working with RNA secondary structure to provide a fast and interactive visualization.Being free of fancy external library dependency and/or network access, the VARNA Applet can be used as a base for a standalone applet. It looks reasonably good and scales up or down nicely to adapt to the space available on a web page, thanks to the anti-aliasing drawing primitives of Swing.
::DEVELOPER
Created by: Kevin Darty, Alain Denise and Yann Ponty ( ponty@lri.fr)
:: SCREENSHOTS
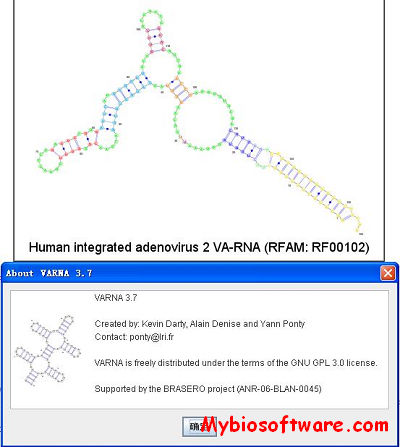
:: REQUIREMENTS
- Windows / MacOsX / Linux
- Java
:: DOWNLOAD
 VARNA
VARNA
:: MORE INFORMATION
Citation
VARNA: Interactive drawing and editing of the RNA secondary structure
Kévin Darty, Alain Denise and Yann Ponty
Bioinformatics, pp. 1974-1975, Vol. 25, no. 15, 2009