proteinhypernetworks 1.0.2
:: DESCRIPTION
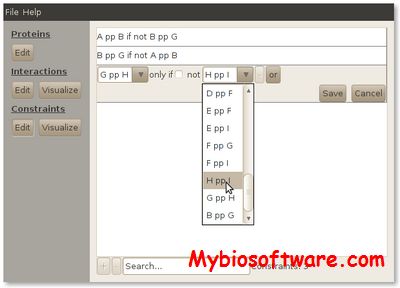
Protein Hypernetworks is a framework for modelling protein networks including boolean interaction dependencies.
::DEVELOPER
Johannes Köster,Eli Zamir (MPI Dortmund), Sven Rahmann
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Johannes Köster, Eli Zamir, Sven Rahmann.
Protein Hypernetworks: a Logic Framework for Interaction Dependencies and Perturbation Effects in Protein Networks;