Bodil 0.8.1
:: DESCRIPTION
Bodil is a modular, multi-platform software package for biomolecular visualization and modeling. Bodil aims to provide easy three-dimensional molecular graphics closely integrated with sequence viewing and sequence alignment editing. Functionality of Bodil is implemented in dynamically loaded modules. Most of the modules available in the present release provide visualization tools, with protein modeling, small-molecule ligand docking and other modules being currently under development. New modules can easily be added via a documented C++ application programming interface.
::DEVELOPER
Structural Bioinformatics Laboratory
:: SCREENSHOTS
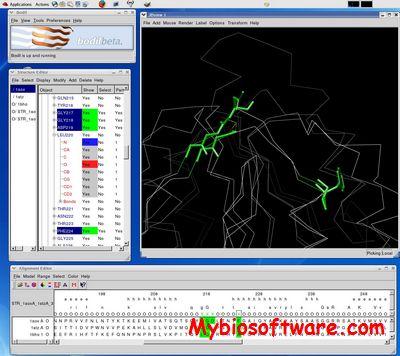
:: REQUIREMENTS
:: DOWNLOAD
 Bodil
Bodil
:: MORE INFORMATION
Citation:
Lehtonen JV, Still DJ, Rantanen VV, Ekholm J, Björklund D, Iftikhar Z, Huhtala M, Repo S, Jussila A, Jaakkola J, Pentikäinen O, Nyrönen T, Salminen T, Gyllenberg M and Johnson M.
BODIL: a molecular modeling environment for structure-function analysis and drug design.
J Comput Aided Mol Des 2004 Jun;18(6):401-419.