YASARA 19.12.14
:: DESCRIPTION
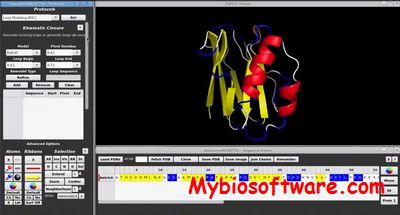
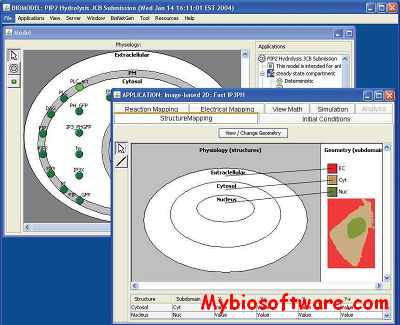
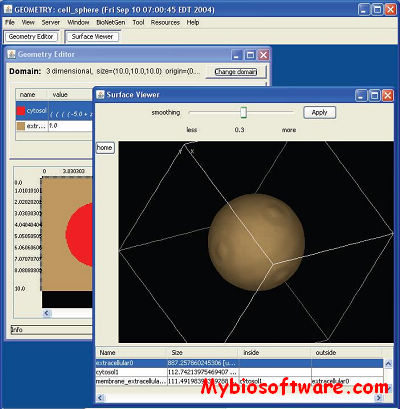
YASARA (Yet Another Scientific Artificial Reality Application)is a molecular-graphics, -modeling and -simulation program.With an intuitive user interface, photorealistic graphics and support for affordable shutter glasses, autostereoscopic displays and input devices, YASARA creates a new level of interaction with the ‘artificial reality’, that allows you to focus on your goal and forget about the details of the program.
YASARA View is available for free and contains the basic functions you need to explore a macromolecular structure interactively, comparable to other molecule viewers.
YASARA Model contains YASARA View and adds all the functions you need to explore, analyze and model small and macromolecules in a production environment. This includes many features you often miss: unlimited undo/redo, macro recorder, quad-buffered stereo with shutter glasses or the DTI virtual window .
YASARA Dynamics contains YASARA Model and adds support for molecular simulations.
YASARA Structure contains YASARA Dynamics and adds all the functions needed to predict and validate macromolecular structures, including ligand docking and highly accurate force fields with knowledge-based potentials, and an optional module for NMR structure determination.
::DEVELOPER
YASARA Biosciences
:: SCREENSHOTS


:: REQUIREMENTS
:: DOWNLOAD
 YASARA
YASARA
:: MORE INFORMATION
Citation
Bioinformatics. 2014 Jul 4. pii: btu426.
YASARA View – molecular graphics for all devices – from smartphones to workstations.
Krieger E1, Vriend G
Bioinformatics. 2002 Feb;18(2):315-8.
Models@Home: distributed computing in bioinformatics using a screensaver based approach.
Krieger E, Vriend G.
 NO
NO