2D-EM
:: DESCRIPTION
2D-EM is a clustering algorithm approach designed for small sample size and high-dimensional datasets. To employ information corresponding to data distribution and facilitate visualization, the sample is folded into its two-dimension (2D) matrix form (or feature matrix). The maximum likelihood estimate is then estimated using a modified expectation-maximization (EM) algorithm.
::DEVELOPER
Laboratory for Medical Science Mathematics
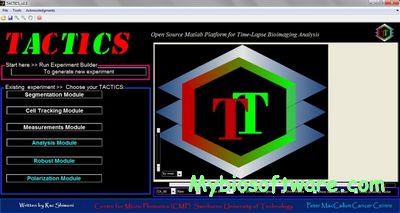
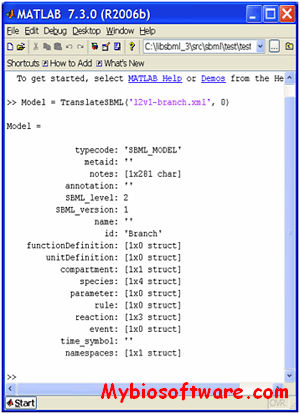
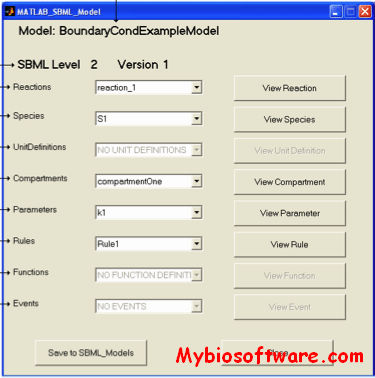
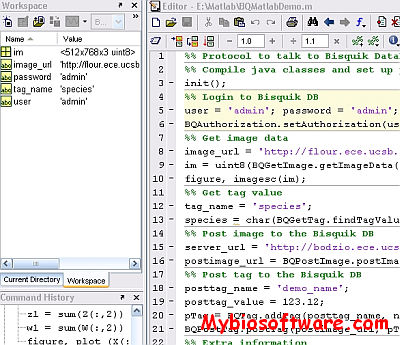
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Matlab
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2017 Dec 28;18(Suppl 16):547. doi: 10.1186/s12859-017-1970-8.
2D-EM clustering approach for high-dimensional data through folding feature vectors.
Sharma A, Kamola PJ, Tsunoda T.