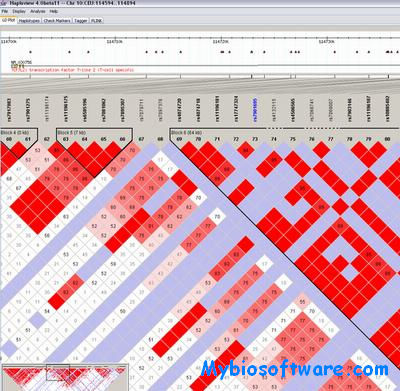
Haploview 4.2
:: DESCRIPTION
Haploview is designed to simplify and expedite the process of haplotype analysis by providing a common interface to several tasks relating to such analyses.
::DEVELOPER
The Analytic and Translational Genetics Unit(AUGT),The Broad Institute,
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Barrett JC, Fry B, Maller J, Daly MJ.
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 2005 Jan 15 [PubMed ID: 15297300]