GMV 0.1
:: DESCRIPTION
The GMV pipeline works as follows. Input is a set of genomes. The pipeline runs PRODIGAL gene predictions on all genomes, runs pan-reciprocal BLAST, and identifies ortholog sets. For a given set of orthologous genes, if the positions of the PRODIGAL selected starts coincide in a multiple sequence alignment, they are accepted. If they do not coincide, a consistent start position is sought where a majority of the highest-scoring PRODIGAL selected sites coincide. If such a position is found, it is accepted, and the predictions are changed for the outlying genes. Otherwise, no start site prediction is made for the ortholog set.
::DEVELOPER
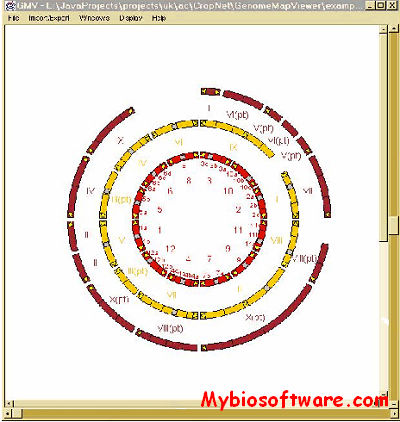
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Genome majority vote improves gene predictions.
Wall ME, Raghavan S, Cohn JD, Dunbar J.
PLoS Comput Biol. 2011 Nov;7(11):e1002284.