LIBFBAT 0.5.1
:: DESCRIPTION
LIBFBAT programming library s robust both to genotyping errors and missing genotypes. The underlying method consists in considering an incomplete data model where the true unobserved genotypes produce the observed one through an explicit genotyping error model.
::DEVELOPER
:: SCREENSHOTS
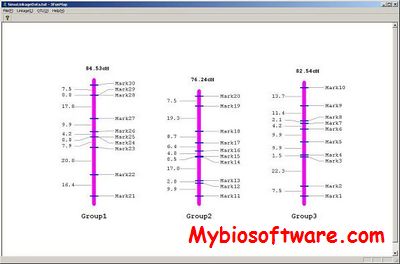
N/A
:: REQUIREMENTS
- Mac OsX/ Linux
- C++ Compiler
:: DOWNLOAD
:: MORE INFORMATION
Citation
Gregory Nuel, Yousri Slaoui and Vincent Miele,
libfbat: a C++ library for family based association testing,
Proceedings of JOBIM, 2008