FBAT v2.05_beta
:: DESCRIPTION
FBAT is an acronym for Family-Based Association Tests in genetic analyses. Family-based association designs, as opposed to case-control study designs, are particularly attractive, since they test for linkage as well as association, avoid spurious associations caused by admixture of populations, and are convenient for investigators interested in refining linkage findings in family samples.
::DEVELOPER
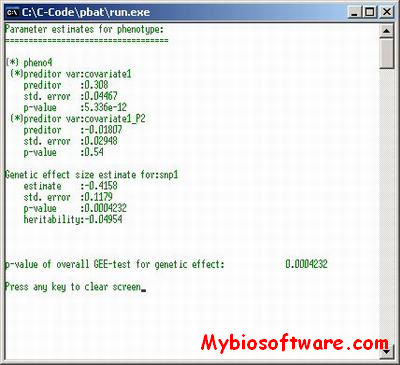
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX /Sun Workstations
:: DOWNLOAD
:: MORE INFORMATION
Citation
Eur J Hum Genet. 2001 Apr;9(4):301-6.
The family based association test method: strategies for studying general genotype–phenotype associations.
Horvath S1, Xu X, Laird NM.