TADA
:: DESCRIPTION
TADA is a novel statistical method that utilizes inherited variation transmitted to affected offspring in conjunction with (de novo) mutations to identify risk genes.
::DEVELOPER
:: SCREENSHOTS
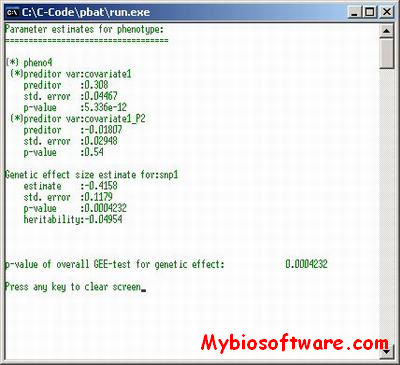
N/A
:: REQUIREMENTS
- Linux/ WIndows/MacOsX
- R package
:: DOWNLOAD
:: MORE INFORMATION
Citation
PLoS Genet. 2013 Aug;9(8):e1003671. doi: 10.1371/journal.pgen.1003671.
Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes.
He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, State MW, Devlin B, Roeder K.