Hybsweeper 02
:: DESCRIPTION
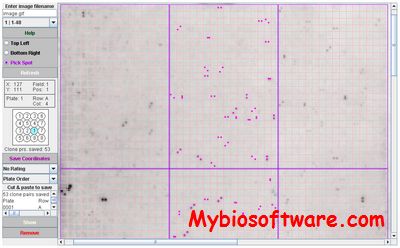
Hybsweeper was developed to handle images of high-density gridding patterns which were matched to clones housed in microtiter dishes. As high-throughput methodologies contribute to large archives of stored clone resources, methods need to be available for convenient processing of hybridization information for the retrieval of the clones needed for research endeavors. A Java software package, Hybsweeper, was developed to select clone identities from autoradiographs which were generated through simple probing methods, overgo probe hybridizations, and isolation of unique clones from saturation hybridization.
::DEVELOPER
Gerard R. Lazo
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/Windows/MacOsX
- Java
:: DOWNLOAD
 Hybsweeper
Hybsweeper
:: MORE INFORMATION
Citation
Gerard R. Lazo, Nancy Lui, Yong Q. Gu, Xiuying Kong, Devin Coleman-Derr, and Olin D. Anderson.
Hybsweeper: a resource for detecting high-density plate gridding coordinates
BioTechniques (2005) 39:320-324.