ClueGO 2.0.5
:: DESCRIPTION
ClueGO is a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network and it can be used in combination with GOlorize. The identifiers can be uploaded from a text file or interactively from a network of Cytoscape. The type of identifiers supported can be easely extended by the user. ClueGO performs single cluster analysis and comparison of clusters. From the ontology sources used, the terms are selected by different filter criteria. The related terms which share similar associated genes can be fused to reduce redundancy. The ClueGO network is created with kappa statistics and reflects the relationships between the terms based on the similarity of their associated genes. On the network, the node colour can be switched between functional groups and clusters distribution. ClueGO charts are underlying the specificity and the common aspects of the biological role.
::DEVELOPER
Integrative Cancer Immunology Laboratory
:: SCREENSHOTS
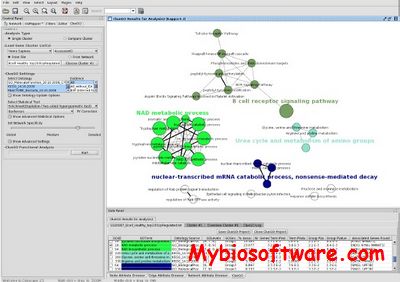
:: REQUIREMENTS
:: DOWNLOAD
 ClueGO
ClueGO
:: MORE INFORMATION
Citation
ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J.
Bioinformatics. 2009 Apr 15;25(8):1091-3. Epub 2009 Feb 23.
 NO
NO