CoNVEM
:: DESCRIPTION
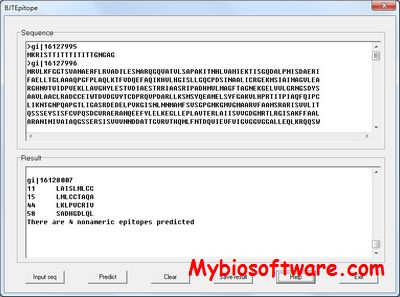
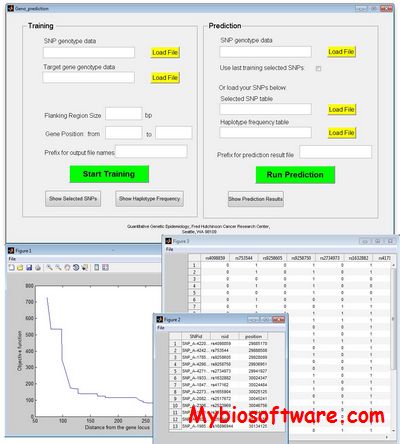
CoNVEM (Copy Number Variation Expectation Maximisation) is an expectation-maximization program for determining allelic spectrum from CNV data (CoNVEM)
::DEVELOPER
Tom Gaunt’s group in the MRC IEU
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation:
Hum Mutat. 2010 Apr;31(4):414-20. doi: 10.1002/humu.21199.
An expectation-maximization program for determining allelic spectrum from CNV data (CoNVEM): insights into population allelic architecture and its mutational history.
Gaunt TR1, Rodriguez S, Guthrie PA, Day IN.