RAP / RapGreen
:: DESCRIPTION
RAP is an algorithm and a software have been developed, which permit to find gene duplications in phylogenetic trees, in order to improve gene function inferences. The algorithm is applicable to realistic data, especially n-ary species tree and unrooted phylogenetic tree
RAP-Green is a new implementation and an improvment of the RAP software which permits to compare gene and species trees, infers duplication events, and provide confidence score in function conservation between genes.
::DEVELOPER
South Green bioinformatics platform
:: SCREENSHOTS
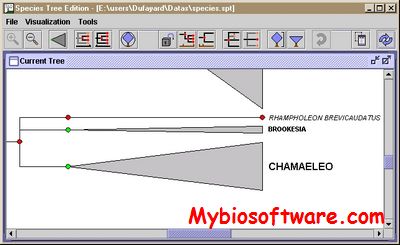
:: REQUIREMENTS
- Linux / Mac OsX / Windows
- Java
:: DOWNLOAD
 RAP , RapGreen
RAP , RapGreen
:: MORE INFORMATION
Citation
Dufayard JF, Bocs S, Guignon V, Larivière D, Louis A, Oubda N, Rouard M, Ruiz M, de Lamotte F.
RapGreen, an interactive software and web package to explore and analyze phylogenetic trees.
NAR Genom Bioinform. 2021 Sep 23;3(3):lqab088. doi: 10.1093/nargab/lqab088. PMID: 34568824; PMCID: PMC8459725.