SEMPHY 2.0 b3
:: DESCRIPTION
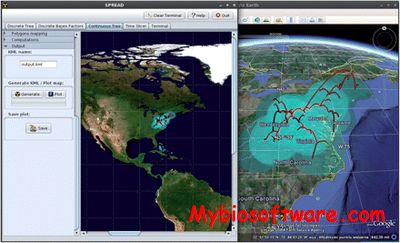
SEMPHY is a tool for data-intensive phylogenetic reconstruction. SEMPHY infers phylogenies by Maximum Likelihood, the most established criterion for finding the correct phylogenetic tree.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/ MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2007 Jan 15;23(2):e136-41.
Phylogeny reconstruction: increasing the accuracy of pairwise distance estimation using Bayesian inference of evolutionary rates.
Ninio M, Privman E, Pupko T, Friedman N.