PubSearch 0.8.1
:: DESCRIPTION
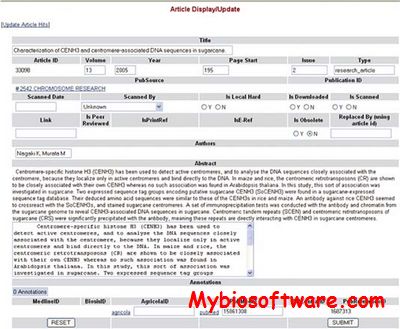
PubSearch is a literature curation management system designed to store and manage the available literature for an organism or system of interest. It provides database curators with a powerful literature search capability, stores relevant biological information, creates automatic associations between the biological information and the literature, and provides a user-friendly web interface for manual validation and curation.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation
Yoo, D, Xu, I, Berardini, T, Rhee, S, Narayanasamy, V, Twigger, S (2006)
PubSearch and PubFetch: a simple management system for semiautomated retrieval and annotation of biological information from the literature..
Curr Protoc Bioinformatics. 2006 Mar;Chapter 9:Unit9.7.