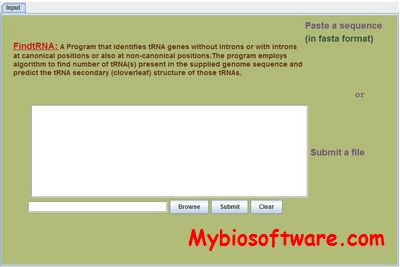
MINTbase V2
:: DESCRIPTION
MINTbase: a web-based framework that serves the dual-purpose of being a content repository for tRFs and a tool for the interactive exploration of these newly discovered molecules.
::DEVELOPER
the Computational Medicine Center at the Sidney Kimmel Medical College of Thomas Jefferson University
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Pliatsika, V, Loher, P, Telonis, AG, Rigoutsos, I.
MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments
Bioinformatics. 2016; In press.