SimMLST 1.0
:: DESCRIPTION
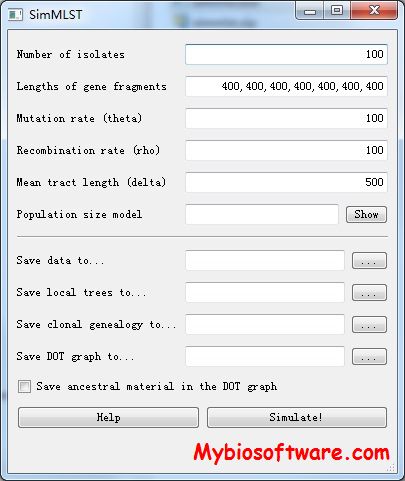
SimMLST, a coalescent method to jointly simulate MLST data and the clonal genealogy that gave rise to the sample. Such simulations are useful to make interpretations about real datasets.Multi-locus sequence typing (MLST) is a widely used method of characterization of bacterial isolates. It has been applied to over 50,000 isolates in over 50 different species and its results are freely available from the pubMLST website.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / MacOsX
- C++ Compiler
:: DOWNLOAD
:: MORE INFORMATION
Citation
Didelot, Lawson and Falush (2009)
SimMLST: simulation of multi-locus sequence typing data under a neutral model
Bioinformatics (2009) 25 (11): 1442-1444.