ATHENA 1.1.0
:: DESCRIPTION
ATHENA (the Analysis Tool for Heritable and Environmental Network Associations) results in clear performance improvements for discovering gene-gene interactions that influence human traits. We employed an alternative tree-based crossover, backpropagation for locally fitting neural network weights, and incorporation of domain knowledge obtainable from publicly accessible biological databases for initializing the search for gene-gene interactions. We tested these modifications in silico using simulated datasets.
::DEVELOPER
:: SCREENSHOTS
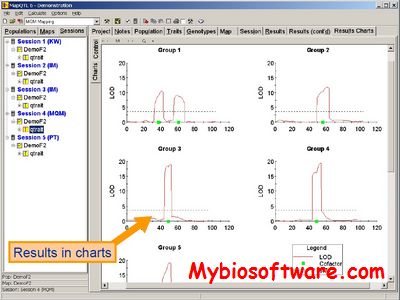
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation:
ATHENA: A knowledge-based hybrid backpropagation-grammatical evolution neural network algorithm for discovering epistasis among quantitative trait Loci.
Turner SD, Dudek SM, Ritchie MD.
BioData Min. 2010 Sep 27;3(1):5.