ModuleAlign
:: DESCRIPTION
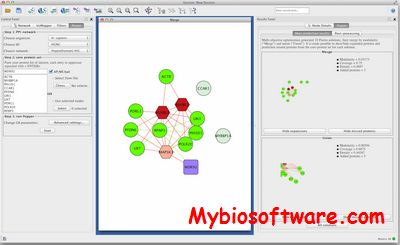
ModuleAlign uses a novel scoring scheme that integrates sequence information and both local and global network topology. It computes a homology score between proteins based on a hierarchical clustering of the input networks. (a) shows the hierarchical structure of a network. Leaves represent the proteins and each cluster joins sets of proteins with similar interactions.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Hashemifar S, Ma J, Naveed H, Canzar S, Xu J.
ModuleAlign: module-based global alignment of protein-protein interaction networks.
Bioinformatics. 2016 Sep 1;32(17):i658-i664. doi: 10.1093/bioinformatics/btw447. PMID: 27587686.