PONDEROSA-CS 2018
:: DESCRIPTION
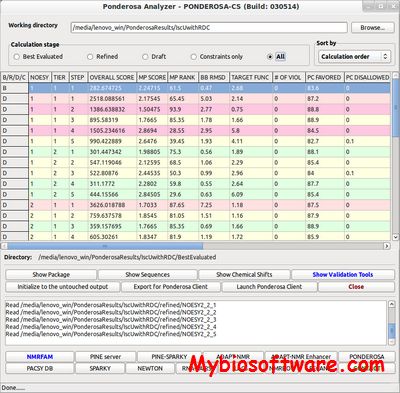
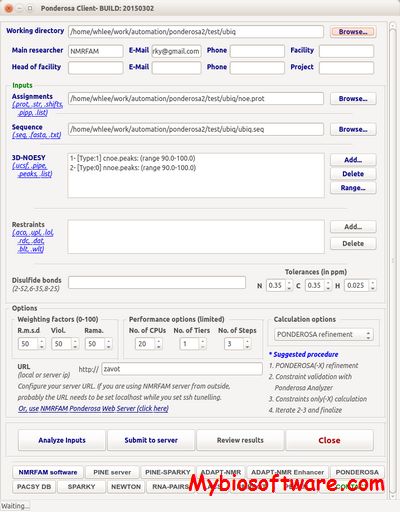
PONDEROSA-CS (Peak-picking Of Noe Data Enabled by Restriction of Shift Assignments with Client/Server ) is a new software package for automated protein 3D structure determination.
::DEVELOPER
NMRFAM (National Magnetic Resonance Facility At Madison)
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Server
:: DOWNLOAD
:: MORE INFORMATION
Citation
J Biomol NMR. 2014 Nov;60(2-3):73-5. doi: 10.1007/s10858-014-9855-x. Epub 2014 Sep 5.
PONDEROSA-C/S: client-server based software package for automated protein 3D structure determination.
Lee W1, Stark JL, Markley JL.
Bioinformatics. 2011 Jun 15;27(12):1727-8. doi: 10.1093/bioinformatics/btr200. Epub 2011 Apr 21.
PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination.
Lee W1, Kim JH, Westler WM, Markley JL.