JAMSS
:: DESCRIPTION
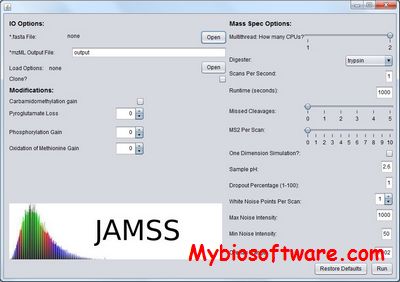
JAMSS is a fast, self-contained in silico simulator capable of generating simulated MS and LC- MS runs while providing meta information on the provenance of each generated signal.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux/ MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2014 Nov 3. pii: btu729.
JAMSS: Proteomics Mass Spectrometry Simulation in Java.
Smith R1, Prince JT