PEPIS 1.0
:: DESCRIPTION
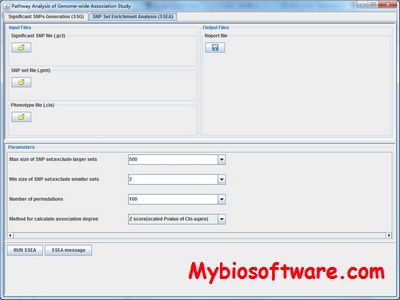
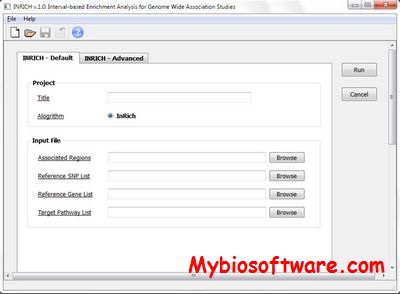
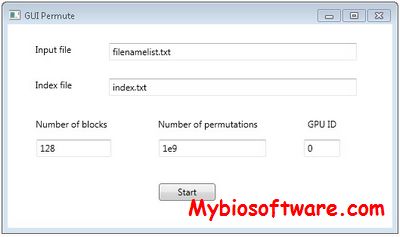
PEPIS is a pipeline that is dedicatedly developed for Polygenic, specifically, Epistatic QTL mapping.
::DEVELOPER
The Zhao Bioinformatics Laboratory
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
PEPIS: A Pipeline for Estimating Epistatic Effects in Quantitative Trait Locus Mapping and Genome-Wide Association Studies.
Zhang W, Dai X, Wang Q, Xu S, Zhao PX.
PLoS Comput Biol. 2016 May 25;12(5):e1004925. doi: 10.1371/journal.pcbi.1004925.