FPC 9.4
:: DESCRIPTION
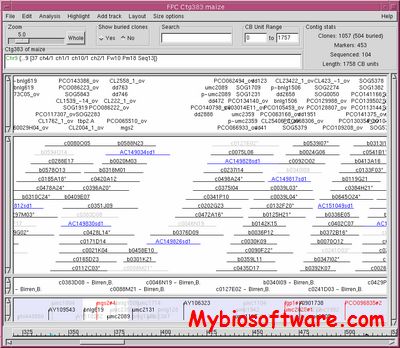
FPC (fingerprinted contigs)is an interactive program for building contigs from fingerprinted clones, where the fingerprint for a clone is a set of restriction fragments.FPC has an algorithm to automatically cluster clones into contigs based on their probability of coincidence score. For each contig, it builds a consensus band (CB) map which is similar to a restriction map but it does not try to resolve all the errors. The CB map is used to assign coordinates to the clones based on their alignment to the map and to provide a detailed visualization of the clone overlap. FPC has editing facilities for the user to refine the coordinates and to remove poorly fingerprinted clones. Functions are available for updating an FPC database with new clones. Contigs can easily be merged, split or deleted. Markers can be added to clones and are displayed with the appropriate contig. Sequence ready clones can be selected and their sequencing status displayed. As such, FPC is an integrated program for the assembly of sequence ready clones for large scale sequencing projects
:: DEVELOPER
Arizona Genomics Computational Lab (AGCoL)
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
 FPC
FPC
:: MORE INFORMATION
Citation:
Nelson, W. and C. Soderlund (2009).
Integrating Sequence with FPC Fingerprinted Maps.
Nucleic Acids Research 1-11 doi:10.1093/nar/gkp034