GenomeComp 1.3
:: DESCRIPTION
GenomeComp is a tool for summarizing, parsing and visualizing the genome wide sequence comparison results derived from voluminous BLAST textual output, so as to locate the rearrangements, insertions or deletions of genome segments between species or strains.
It can be easily used to compare, parsing and visualize large genomic sequences, especially closely related genomes such as inter-species or inter-strains. In addition, it can also show other sequence features like repeat sequence distributions in one whole-genome DNA sequence by comparing the genome to itself.
::DEVELOPER
Chinese National Human Genome Center
:: SCREENSHOTS
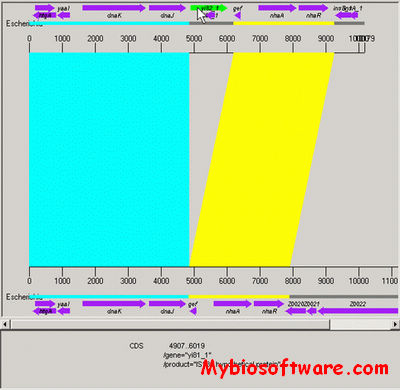
:: REQUIREMENTS
- Windows / Linux / Solaris / IRIX
:: DOWNLOAD
 GenomeComp
GenomeComp
:: MORE INFORMATION
Citation
Yang, J., Wang, J.H., Yao, Z.J., Jin, Q., Shen, Y., Chen, R.S., 2003.
GenomeComp: a visualization tool for microbial genome comparison.
J. Microbiol. Meth. 54, 423-426.