BEAST 1.10.4 / BEAST2 2.6.1
:: DESCRIPTION
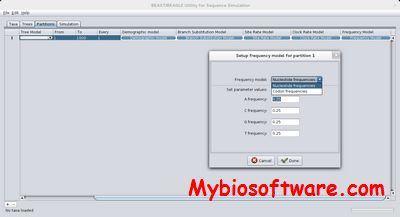
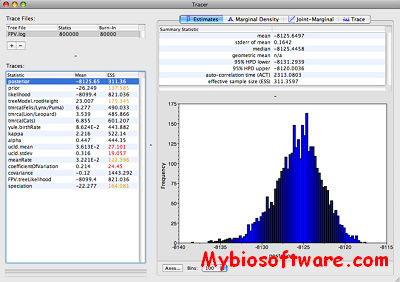
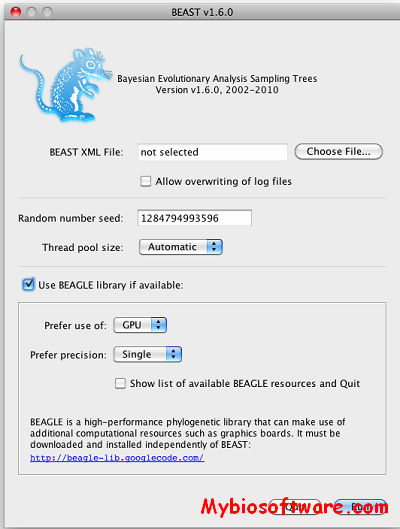
BEAST (Bayesian Evolutionary Analysis Samling Trees) is a cross-platform program for Bayesian MCMC analysis of molecular sequences. It is entirely orientated towards rooted, time-measured phylogenies inferred using strict or relaxed molecular clock models. It can be used as a method of reconstructing phylogenies but is also a framework for testing evolutionary hypotheses without conditioning on a single tree topology. BEAST uses MCMC to average over tree space, so that each tree is weighted proportional to its posterior probability. We include a simple to use user-interface program for setting up standard analyses and a suit of programs for analysing the results.
BEAST 2 is an open source cross-platform program for Bayesian MCMC phylogenetic analysis of molecular sequences.
::DEVELOPER
The University of Auckland Computational Evolution Group
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOS
- Java
:: DOWNLOAD
 BEAST /BEAST2
BEAST /BEAST2
:: MORE INFORMATION
Citation:
BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis.
Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ.
PLoS Comput Biol. 2019 Apr 8;15(4):e1006650. doi: 10.1371/journal.pcbi.1006650.
Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration.
Gavryushkina A, Welch D, Stadler T, Drummond AJ.
PLoS Comput Biol. 2014 Dec 4;10(12):e1003919. doi: 10.1371/journal.pcbi.1003919.
Alexei J Drummond and Andrew Rambaut
BEAST: Bayesian evolutionary analysis by sampling trees
BMC Evolutionary Biology 2007, 7:214doi:10.1186/1471-2148-7-214