BayesDiallel 0.982
:: DESCRIPTION
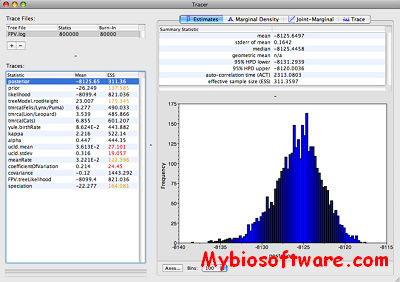
BayesDiallel is a Gibbs-sampler program designed to fit, image, and give confidence for intricately modeled inheritance in Diallel F1 cross data.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux /MacOsX / Windows
- R package
:: DOWNLOAD
:: MORE INFORMATION
Citation
Lenaric A, Svenson K, Churchill G, Valdar W, (2012)
A General Bayesian Approach to analyzing Diallel Crosses of Infred Strains
Genetics, Vol. 190, 413-435, February 2012.