CellC 1.2
:: DESCRIPTION
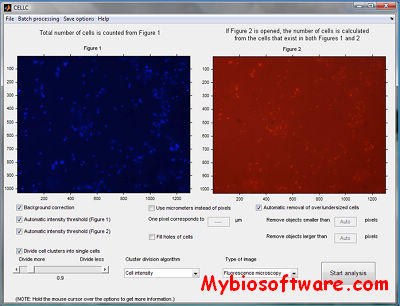
CellC (Cell Counting) was developed and validated for quantification of bacterial cells from digital microscope images. CellC enables automated enumeration of bacterial cells, comparison of total count and specific count images [e.g., 4′,6-diamino-2-phenylindole (DAPI) and fluorescence in situ hybridization (FISH) images], and provides quantitative estimates of cell morphology. The software includes an intuitive graphical user interface that enables easy usage as well as sequential analysis of multiple images without user intervention. Validation of enumeration reveals correlation to be better than 0.98 when total bacterial counts by CellC are compared with manual enumeration, with all validated image types.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- MATLAB
- Image Processing Toolbox 5.0 or higher
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Jyrki Selinummi, Jenni Seppälä, Olli Yli-Harja, and Jaakko A. Puhakka,
Software for quantification of labeled bacteria from digital microscope images by automated image analysis
BioTechniques, Volume 39, Number 6: pp 859-863.