mswordtree 1.0
:: DESCRIPTION
mswordtree is a tool for generating graphical phylogenetic trees in a Microsoft Word document directly from a Newick format description. The resulting graphic is a Word picture and so can then be moved easily between MS Office applications and extensively edited. mswordtree is a Word Basic macro and can therefore be integrated directly into your installation of Word, eliminating the need for an external program to graphically render your trees.
::DEVELOPER
DICKS COMPUTATIONAL BIOLOGY GROUP
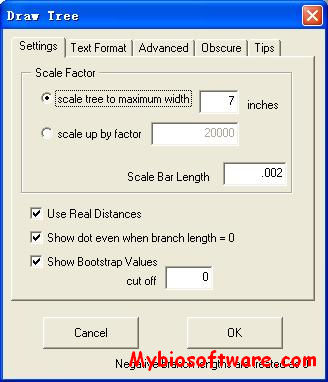
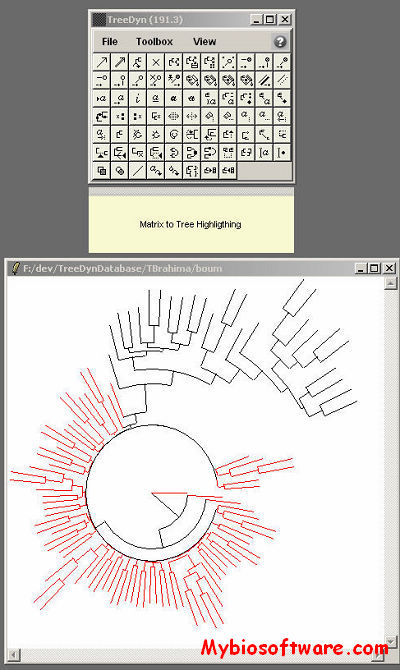
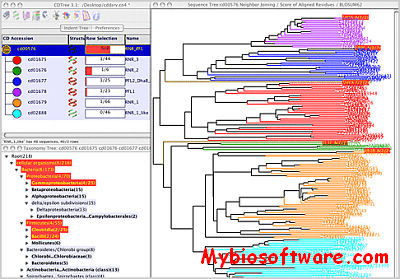
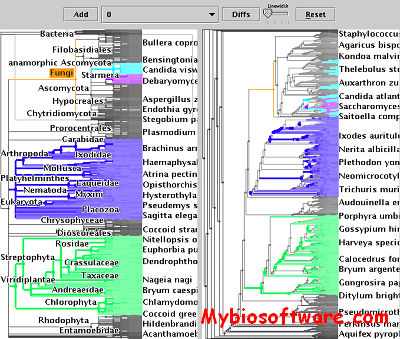
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Mac OsX
- MS Word
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Savva G, Conn J and Dicks J (2004)
Drawing phylogenetic trees in LaTeX and Microsoft Word
Bioinformatics 20(14): 2322-2323.