BioCham 4.5.17
:: DESCRIPTION
BioCham (Biochemical Abstract Machine) is a modelling environment for systems biology, with some unique features for inferring unknown model parameters from temporal logic constraints.
BioCham is mainly composed of :
Advertisement
- a rule-based language for modeling biochemical systems (compatible with SBML)
- several simulators (boolean, differential, stochastic),
- a temporal logic based language to formalize the temporal properties of a biological system and validate models with respect to such specifications,
- unique features for developing/correcting/completing/coupling models, including the inference of kinetic parameters in high dimension from temporal logic constraints.
::DEVELOPER
The “Contraintes” project-team
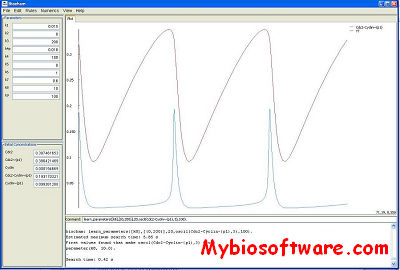
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux / Mac OsX
- JAVA
- this patched version (including source and binary versions) of the model checker NuSMV
- GNU Plot
- Graphviz
:: DOWNLOAD
:: MORE INFORMATION
Citation
ioinformatics. 2006 Jul 15;22(14):1805-7. Epub 2006 May 3.
BIOCHAM: an environment for modeling biological systems and formalizing experimental knowledge.
Calzone L, Fages F, Soliman S.