APID2NET
:: DESCRIPTION
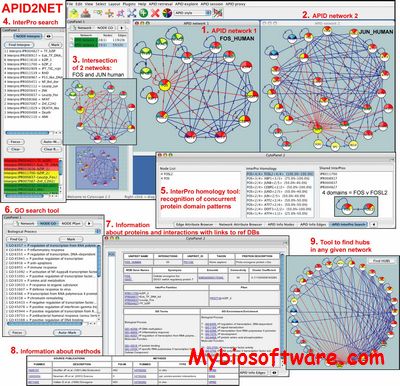
APID2NET is an open access tool, included in Cytoscape, that allows to surf unified interactome data by quering APID server and facilitates dynamic analysis of the protein-protein interaction (PPI) networks. The program is designed to visualize, dynamically explore and analyze the proteins and interactions retrieved, including all the annotations and attributes associated to such PPIs, i.e.: GO terms, Pfam and InterPro domains, experimental methods that validate each interaction, PubMed IDs, UniProt IDs, etc. The tool provides a rich interactive graphical representation of the networks with all Cytoscape capabilities, plus new automatic tools to find and locate concurrent functional and structural attributes along all protein pairs in a network. A hubs location tool is also implemented.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation
Hernandez-Toro J., Prieto C. and De Las Rivas J. (2007).
APID2NET: unified interactome graphic analyzer.
Bioinformatics 23(18): 2495-2497. PMID: 17644818.