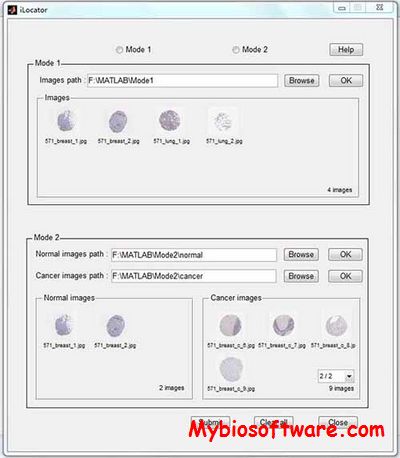
Cell-PLoc 2.0
:: DESCRIPTION
Cell-PLoc is a package of Web servers for predicting subcellular localization of proteins in various organisms.The package contains the following six predictors: Euk-mPLoc 2.0 , Hum-mPLoc 2.0, Plant-PLoc, Gpos-PLoc, Gneg-PLoc and Virus-PLoc, specialized for eukaryotic, human, plant, Gram-positive bacterial, Gram-negative bacterial and viral proteins, respectively.
::DEVELOPER
Computational Systems Biology Group, Shanghai Jiao Tong University
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation:
Nat Protoc. 2008;3(2):153-62. doi: 10.1038/nprot.2007.494.
Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms.
Chou KC1, Shen HB.
Kuo-Chen Chou, and Hong-Bin Shen,
Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms,
Natural Science, 2010, 2: 1090-1103
PLoS One. 2010 Apr 1;5(4):e9931. doi: 10.1371/journal.pone.0009931.
A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0.
Chou KC1, Shen HB.
A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0.
Shen HB, Chou KC.
Anal Biochem. 2009 Nov 15;394(2):269-74. doi: 10.1016/j.ab.2009.07.046.
Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization.
Chou KC, Shen HB.
PLoS One. 2010 Jun 28;5(6):e11335. doi: 10.1371/journal.pone.0011335.
Gpos-mPLoc: a top-down approach to improve the quality of predicting subcellular localization of Gram-positive bacterial proteins.
Shen HB, Chou KC.
Protein Pept Lett. 2009;16(12):1478-84.
Gneg-mPLoc: a top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins.
Shen HB, Chou KC.
J Theor Biol. 2010 May 21;264(2):326-33. doi: 10.1016/j.jtbi.2010.01.018.
J Biomol Struct Dyn. 2010 Oct;28(2):175-86.
Virus-mPLoc: a fusion classifier for viral protein subcellular location prediction by incorporating multiple sites.
Shen HB1, Chou KC.