AffyMAPSDetector 1.0
:: DESCRIPTION
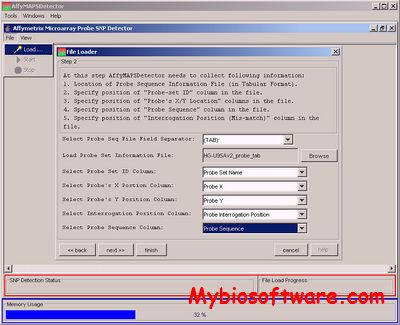
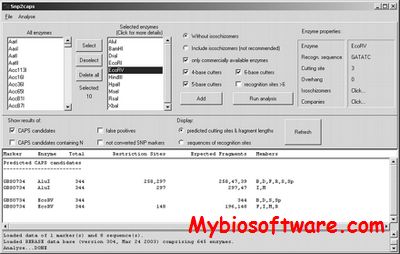
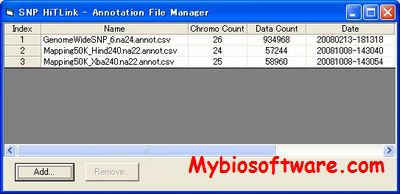
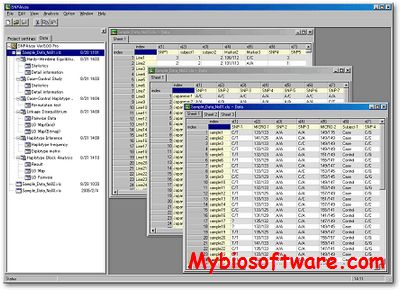
AffyMAPSDetector stands for Affymetrix MicroArray Probe SNP Detector. It detects the presence of SNPs (Single Nucleotide Polymorphism) in the probes of Affymetrix GeneChipTM expression arrays. However, it can be used to characterize any GeneChipTM with respect to SNPs for which NCBI fully supports gene and SNP annotations (i.e. the organism for which GeneChipTM is desinged and that organism’s genome exists in NCBI nucleotide and SNP database).
DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2007 Jul 30;8:276.
AffyMAPSDetector: a software tool to characterize Affymetrix GeneChip expression arrays with respect to SNPs.
Kumari S, Verma LK, Weller JW.