SDT v1.2
:: DESCRIPTION
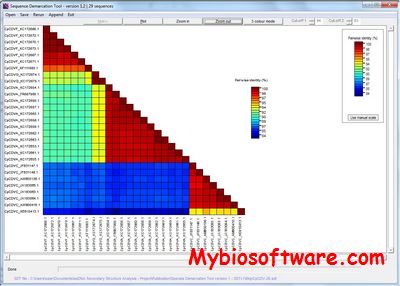
SDT is a free and easy to use program that allows classification of virus sequences based on sequence pairwise identity. It takes as input a FASTA file of aligned or unaligned DNA or protein sequences and aligns every unique pair of sequences, calculates pairwise similarity scores, and displays a colour coded matrix of these scores. It also produces both a plot of these pairwise identity scores and text files containing analysis results.
::DEVELOPER
the University of Cape Town (UCT) Computational Biology (CBIO) Group
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation:
PLoS One. 2014 Sep 26;9(9):e108277. doi: 10.1371/journal.pone.0108277. eCollection 2014.
SDT: a virus classification tool based on pairwise sequence alignment and identity calculation.
Muhire BM1, Varsani A2, Martin DP