NEMO v0.1
:: DESCRIPTION
NEMO is a multi-omic clustering algorithm. NEMO supports data in which some omics were measured for only a subset of the samples.
::DEVELOPER
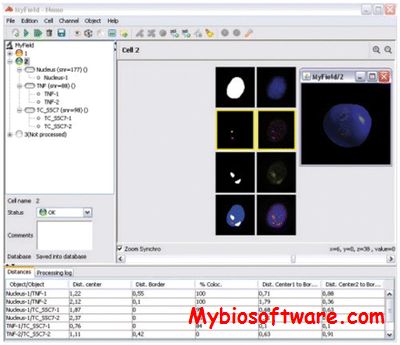
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/ MacOsX
- R
- SNFtool
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2019 Sep 15;35(18):3348-3356. doi: 10.1093/bioinformatics/btz058.
NEMO: cancer subtyping by integration of partial multi-omic data.
Rappoport N, Shamir R.